CytoGnomix’s Mutation Forecaster: Key to discovery of mutations in novel ALS gene. Nicolas et al. Genome-wide Analyses Identify KIF5A as a Novel ALS Gene. Neuron 97:1268-1283.e6, 2018
http://www.cell.com/neuron/fulltext/S0896-6273(18)30148-X
From our subscriber, Dr. John Landers (U. Mass. School of Medicine):
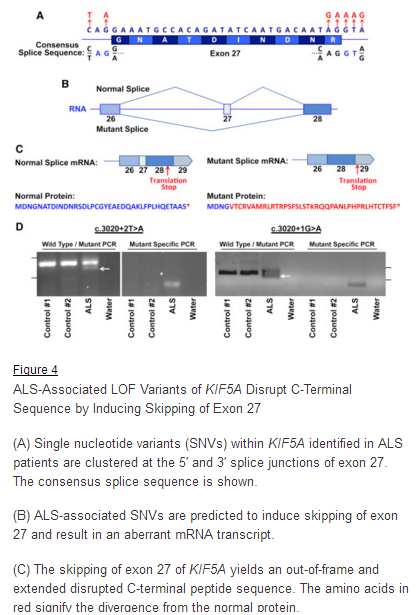
“We used the application ASSEDA (Automated Splice Site and Exon Definition Analyses) to predict any mutant mRNA splice isoforms resulting from these variants (Mucaki et al. 2013). This algorithm was chosen as it is known to have high performance in splice prediction (Caminsky et al., 2014). ASSEDA predicted a complete skipping of exon 27 for all variants, yielding a transcript with a frameshift at coding amino acid 998, the deletion of the normal C-terminal 34 amino acids of the cargo-binding domain, and the extension of an aberrant 39 amino acids to the C terminus (Table 3; Figures 4B and 4C). The presence of transcripts with skipped exon 27 was demonstrated by performing…”