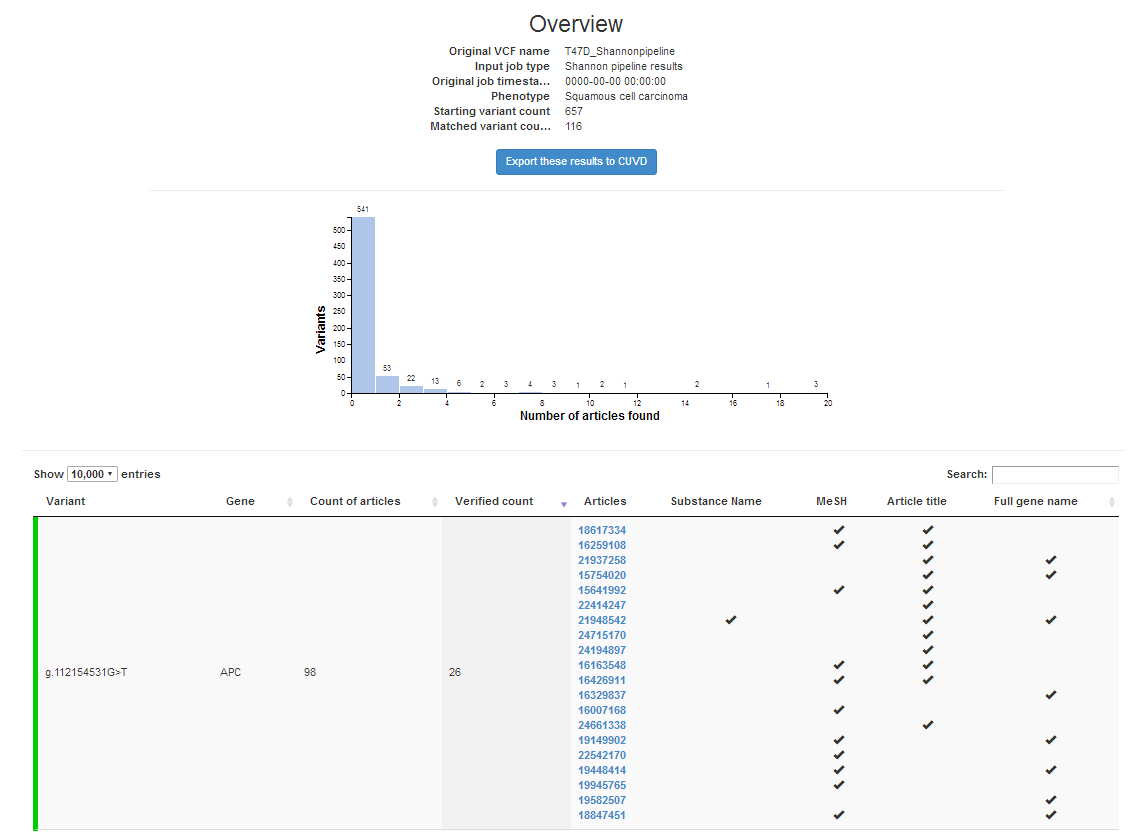
We have just updated the CytoVA software product. It now contains all PubMed references through Dec 2018 and the most recent version of the human phenotype ontology (HPO). With CytoVA (see below), you can query Shannon pipeline, Veridical or VEP output, the list of predicted deleterious variants, with this software for HPO clinical phenotypes. The software reports the articles, and PubMed index codes that support the relationship between the gene variants and the queried phenotype. It’s a very effective way of filtering exome or complete genome sequencing results to limit the best candidate variants.