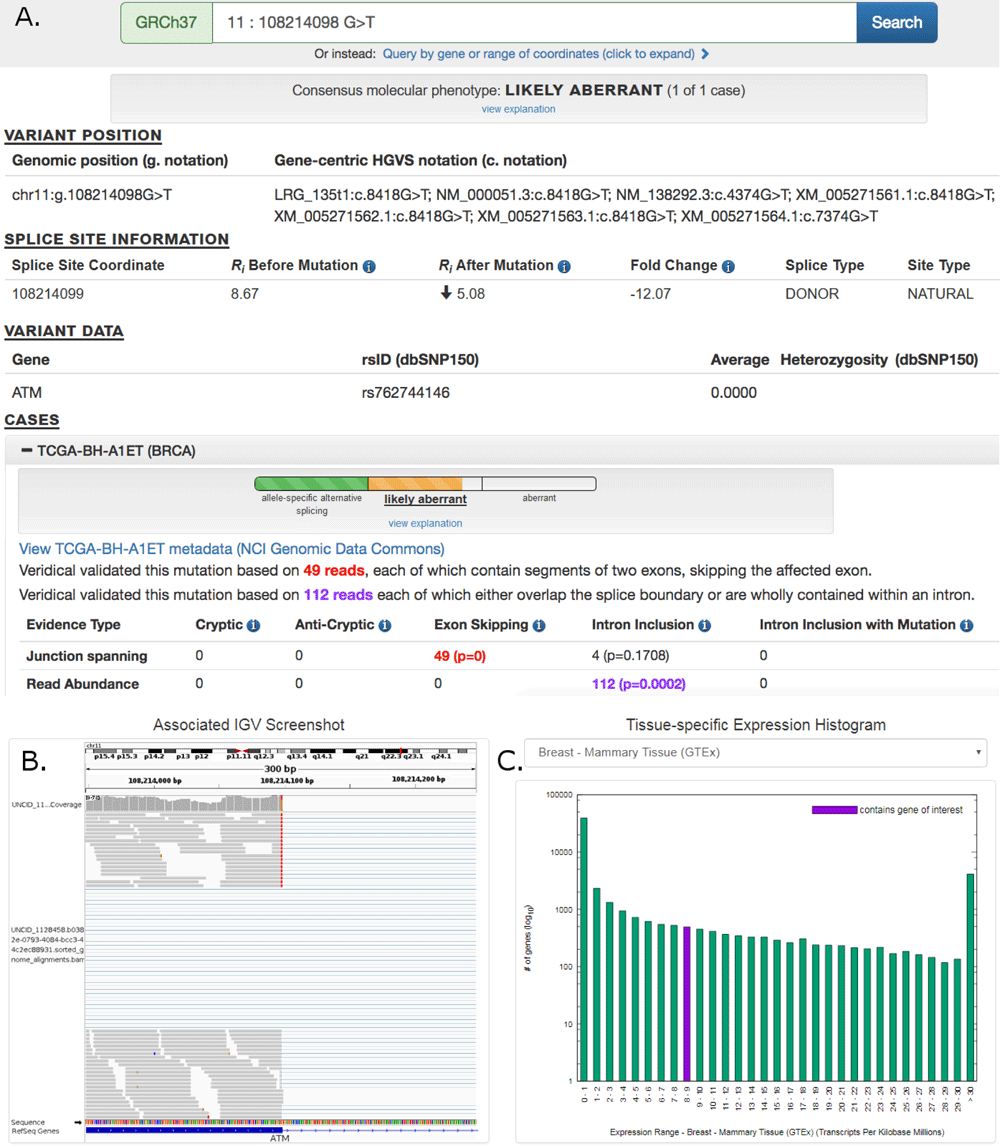
We have published another revision to our pan-cancer splicing mutation database paper: Shirley BC, Mucaki EJ and Rogan PK. Pan-cancer repository of validated natural and cryptic mRNA splicing mutations F1000Research 2019, 7:1908 (https://f1000research.
In this version, we derive a simplified variant classification scheme with ClinVar designations calibrated to the molecular phenotypes of mRNA splicing mutations. These are now indicated in the ValidspliceMut (https://validsplicemut.