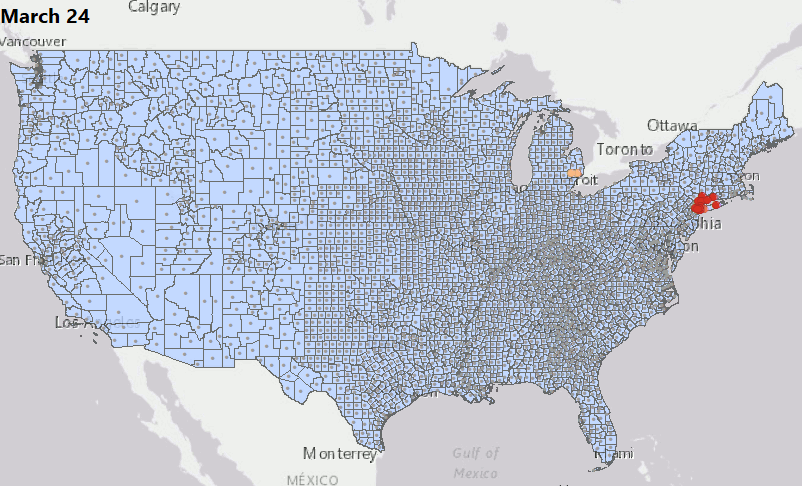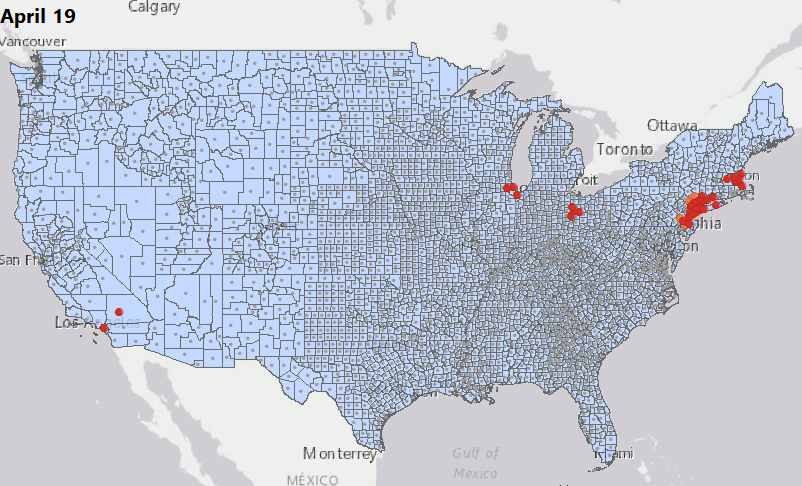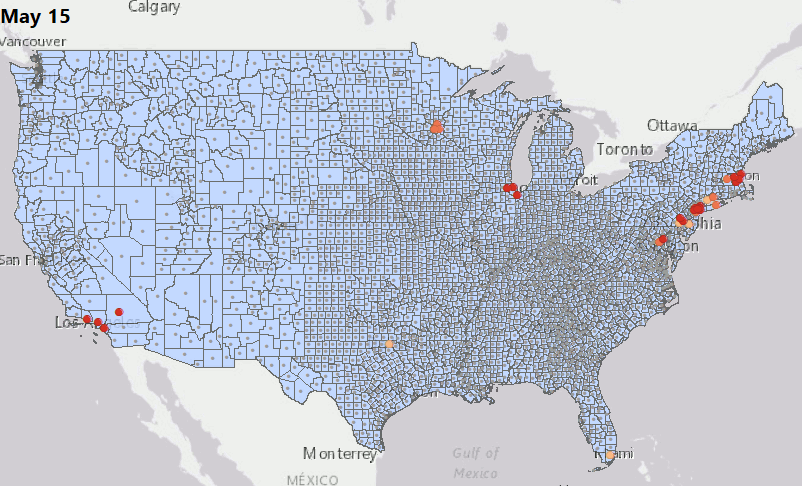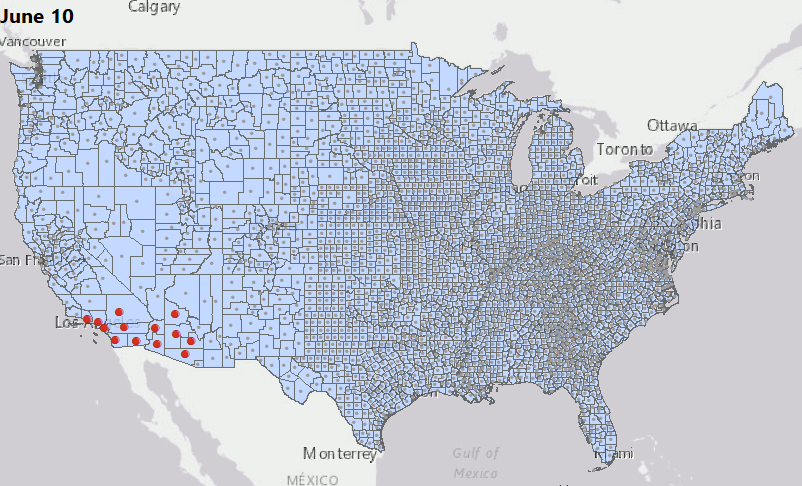
We have consolidated our geostatistical biodosimetry contributions at Grow Kudos. We have been developing a new application of this technology for tracking hotspots of infections during the current COVID-19 pandemic. This animation shows geostatistically significant hotspot counties in the US relative to 3 nearest neighbors from March through June 2020. Please stay tuned for more:
April 24, 2020. Schulich School of Medicine and Dentistry highlights PLOS ONE study on radiation dose estimation and COVID19
Rogan PK, Mucaki EJ, Lu R, Shirley BC, Waller E, Knoll JHM (2020) Meeting radiation dosimetry capacity requirements of population-scale exposures by geostatistical sampling. PLoS ONE 15(4): e0232008. https://doi.org/10.1371/journal.pone.0232008
April 24, 2020. Assessing radiation exposure across a population by geolocation
Feb. 11, 2020. Article about allele-specific alternative splicing published
Mucaki, E.J., Shirley, B.C. and Rogan, P.K., 2020. Expression changes confirm genomic variants predicted to result in allele-specific, alternative mRNA splicing. Frontiers in Genetics, 11: 109 (https://www.frontiersin.org/articles/10.3389/fgene.2020.00109/full).
(preprint in bioRxiv, 549089 [doi: https://doi.org/10.1101/549089])
Nov. 18, 2019. Final version of review article on prediction of chemotherapy response by machine learning now published
Multigene signatures of responses to chemotherapy derived by biochemically-inspired machine learning
Journal:Molecular Genetics and Metabolism
Volume 128, Issues 1–2, September–October 2019, Pages 45-52
PK Rogan.
Sept. 7, 2019. Pan cancer article and database update
Aug. 19, 2019. Review article about machine learning for predicting chemotherapy response
New review article in Molecular Genetics and Metabolism about predicting responses to chemotherapy:
Multigene signatures of responses to chemotherapy derived by biochemically inspired machine learning
Published: https://doi.org/10.1016/j.ymgme.2019.08.005
Abstract: Pharmacogenomic responses to chemotherapy drugs can be modeled by supervised machine learning of expression and copy number of relevant gene combinations. Such biochemical evidence can form the basis of derived gene signatures using cell line data, which can subsequently be examined in patients that have been treated with the same drugs. These gene signatures typically contain elements of multiple biochemical pathways which together comprise multiple origins of drug resistance or sensitivity. The signatures can capture variation in these responses to the same drug among different patients.
July 31, 2019. New preprint on biodosimetry in BioRxiv
“Automated Cytogenetic Biodosimetry at Population-Scale”
, , , , , , , , , , , , ,
, ,
https://www.biorxiv.org/content/10.1101/718973v1 (doi: https://doi.org/10.1101/718973)
Introduction The dicentric chromosome (DC) assay accurately quantifies exposure to radiation, however manual and semi-automated assignment of DCs has limited its use for a potential large-scale radiation incident. The Automated Dicentric Chromosome Identifier and Dose Estimator Chromosome (ADCI) software automates unattended DC detection and determines radiation exposures, fulfilling IAEA criteria for triage biodosimetry. We present high performance ADCI (ADCI-HT), with the requisite throughput to stratify exposures of populations in large scale radiation events.
Methods ADCI-HT streamlines dose estimation by optimal scheduling of DC detection, given that the numbers of samples and metaphase cell images in each sample vary. A supercomputer analyzes these data in parallel, with each processor handling a single image at a time. Processor resources are managed hierarchically to maximize a constant stream of sample and image analysis. Metaphase data from populations of individuals with clinically relevant radiation exposures after simulated large nuclear incidents were analyzed. Sample counts were derived from US Census data. Analysis times and exposures were quantified for 15 different scenarios.
Results Processing of metaphase images from 1,744 samples (500 images each) used 16,384 CPUs and was completed in 1hr 11min 23sec, with radiation dose of all samples determined in 32 sec with 1,024 CPUs. Processing of 40,000 samples with varying numbers of metaphase cells, 10 different exposures from 5 different biodosimetry labs met IAEA accuracy criteria (dose estimate differences were < 0.5 Gy; median = 0.07) and was completed in ~25 hours. Population-scale metaphase image datasets within radiation contours of nuclear incidents were defined by exposure levels (either >1 Gy or >2 Gy). The time needed to analyze samples of all individuals receiving exposures from a high yield airborne nuclear device ranged from 0.6-7.4 days, depending on the population density.
Conclusion ADCI-HT delivers timely and accurate dose estimates in a simulated population-scale radiation incident.
July 4, 2019. Presentations describing interlaboratory comparison of radiation exposure determination by automated cytogenetic biodosimetry
We will be presenting:
Determination of radiation exposure levels by fully automated
dicentric chromosome analysis: Results from IAEA MEDBIODOSE
(CRP E35010) interlaboratory comparison
at both the 19th International Congress of Radiation Research (Aug. 25-29, 2019) and the 12th International Symposium on Chromosome Aberrations (Aug. 27, 2019) in Manchester, UK. This study compared the performance of our Automated Dicentric Chromosome Identifier and Dose Estimator (ADCI) using data from 6 different laboratories. Each of these members of the IAEA-sponsored Cooperative Research Project E35010, submitted images for calibration curve construction and at least 2 samples of unknown exposure to CytoGnomix for analysis with ADCI. We will report the results of this analysis during this presentation.
This poster presentation is now available on the Zenodo website (http://doi.org/10.5281/zenodo.4012749)
doi: DOI 10.5281/zenodo.4012748
Authors:
Rogan P , Shirley B , Li Y , Guogyte K , Sevriukova O , Ngoc Duy P , Moquet J ,
Ainsbury E , Sudprasert W , Wilkins R , Norton F , Knoll J
Department of Biochemistry , University of Western Ontario, London Ontario, Canada
Department of Pathology and Laboratory Medicine, University of Western Ontario, London
Ontario, Canada
Radiation Protection Centre, Ministry of Health (L T -RPC), Vilnius, Lithuania
Dalat Nuclear Research Institute (VN-DNRI), Dalat, Vietnam
Public Health England (PHE), Oxford, Great Britain
Thai Biodosimetry Network, Kasetsart University (THA), Bangkok, Thailand
Health Canada, Ottawa Ontario, Canada
Canadian Nuclear Laboratories, Chalk River Ontario, Canada
Cytognomix, London Ontario, Canada
June 28, 2019. Platform Presentation at 2019 ICRR on population-scale biodosimetry
We will present a new geostatistical approach to reduce biodosimetry workload in a large scale nuclear event at the International Congress of Radiation Research in Manchester UK, 25-20 August, 2019:
(link to full abstract in conference program)
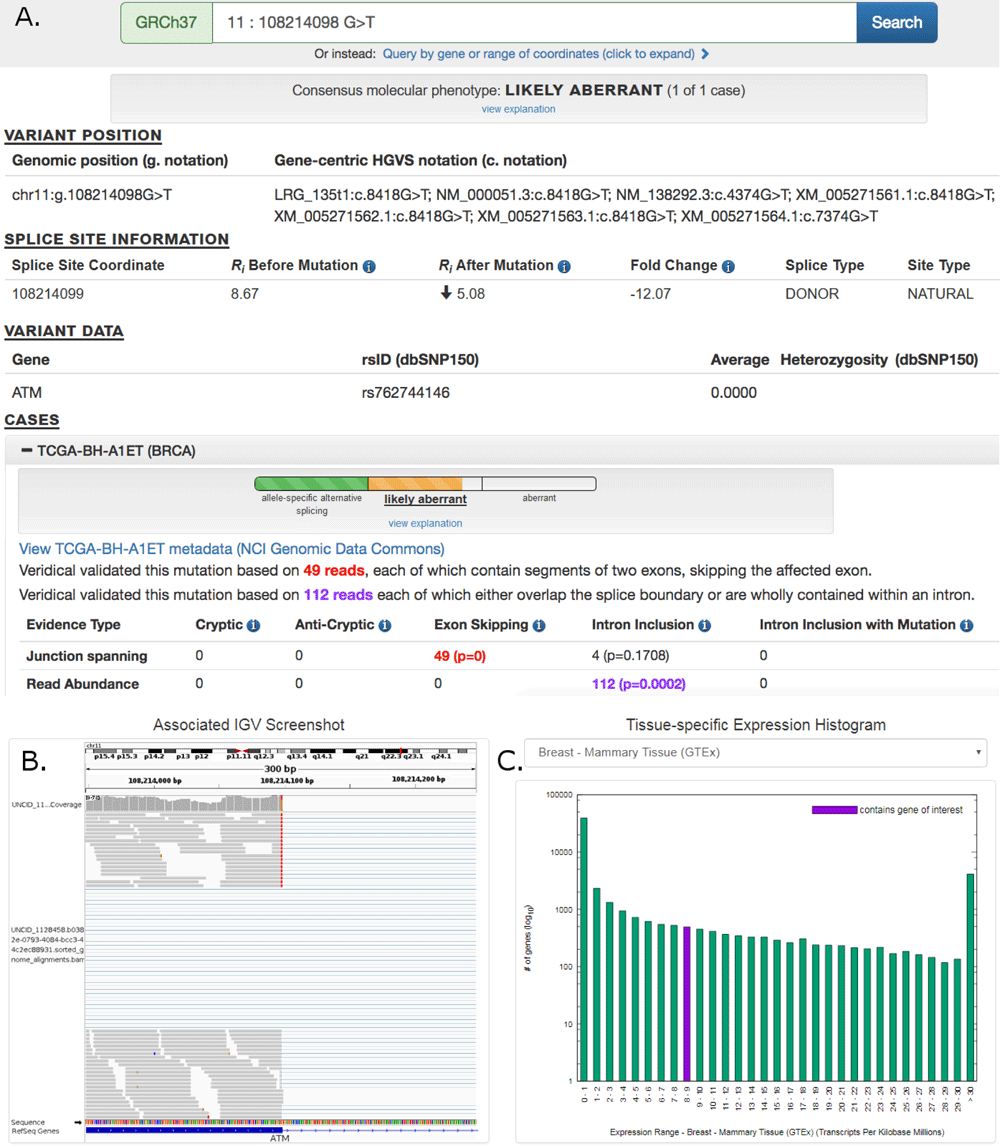
May 30, 2019. F1000Research splicing mutation database article now indexed in PubMed
Large repository of human mRNA splicing mutations in TCGA and ICGC tumor exomes and genomes, with every mutation validated by transcriptome analysis of tumor vs controls:
Shirley BC, Mucaki EJ and Rogan PK. Pan-cancer repository of validated natural and cryptic mRNA splicing mutations. F1000Research 2019, 7:1908 (https://f1000research.com/articles/7-1908/v2)
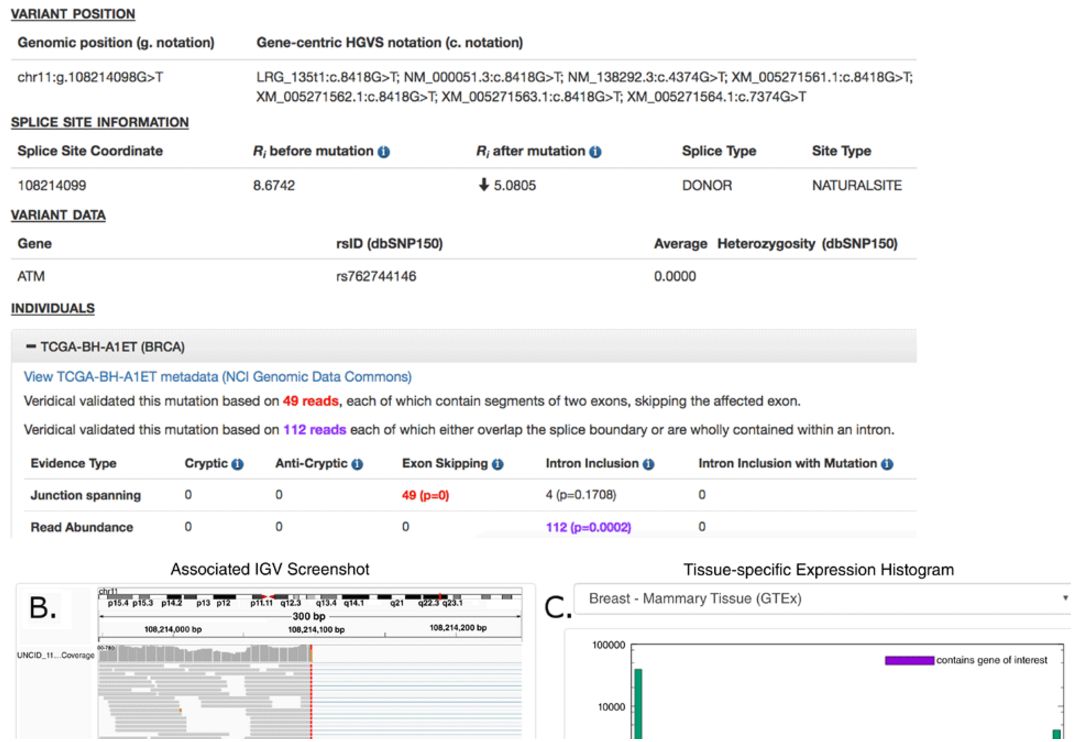
April 11, 2019. New article about transcription factor binding site clusters, their role in target gene regulation and mutation analysis
This article has now been accepted for publication and indexing in PubMed:
Lu R and Rogan PK. Transcription factor binding site clusters identify target genes with similar tissue-wide expression and buffer against mutations [version 2; peer review: 2 approved]. F1000Research 2019, 7:1933 (https://doi.org/10.12688/f1000research.17363.2)
Pan-Cancer Splicing Web Beacon to be Presented at ACMG Meeting
|
|
|
March 20, 2019. Presentation at the 2019 American College of Medical Genetics and Genomics annual conference
The following paper has been accepted for presentation:
“Pan-Cancer Repository of Validated Natural and Cryptic mRNA Splicing Mutations”,
Category: “Laboratory genetics and genomics”, Abstract Poster Number: 754 (link to Abstract)
Where: Exhibit hall, Washington Convention Center, ACMG Clinical Genetics Meeting in Seattle, Washington
When: April 2 – 6, 2019; Poster presentation time: Friday, 4/5 from 10:30am-12:00pm
This work will be available as an ePoster AS WELL AS being presented in printed format on a poster board during the Annual Meeting. Details to access the ePoster will be available soon.
Feb. 14, 2019. New article about targeted validation of mRNA splicing mutations
New article in bioRxiv:
Expression changes confirm predicted single nucleotide variants affecting mRNA splicing. E. J. Mucaki and P.K. Rogan. (https://www.biorxiv.org/content/10.1101/549089v1)
This paper describes high quality qRT-PCR and microarray expression data of predicted splicing variants. The results confirm results of genome-wide TCGA and ICGC RNASeq findings (https://f1000research.com/articles/7-1908/v1). The genome scale results were obtained using CytoGnomix’s MutationForecaster (http://mutationforecaster.com ) system.
Jan. 2019. CytoGnomix licenses ADCI software for radiation biodosimetry
CytoGnomix is licensing its Automated Dicentric Chromosome Identifier and Dose Estimator product to a Canadian government-owned contractor-operated laboratory through 2020. This organization’s primary business is in nuclear science and technology.
January 24, 2019. Updated version of CytoGnomix Visual Analytics (CytoVA) on MutationForecaster
We have just updated the CytoVA software product. It now contains all PubMed references through Dec 2018 and the most recent version of the human phenotype ontology (HPO). With CytoVA (see below), you can query Shannon pipeline, Veridical or VEP output, the list of predicted deleterious variants, with this software for HPO clinical phenotypes. The software reports the articles, and PubMed index codes that support the relationship between the gene variants and the queried phenotype. It’s a very effective way of filtering exome or complete genome sequencing results to limit the best candidate variants.
January 9, 2019. Article on fully automated interpretation of the dicentric chromosome assay for radiation quantification now available
Our paper, “RADIATION DOSE ESTIMATION BY COMPLETELY AUTOMATED INTERPRETATION OF THE DICENTRIC CHROMOSOME ASSAY” is now published in the journal Radiation Protection Dosimetry.
Unfortunately, the journal has not made the article open access. We have made it available on our ADCIWiki website, as permitted by the copyright agreement.
The link to the pdf full text is: