Author Archives: Peter Rogan
September 7, 2015. Video presentation of Molecular Oncology article on chemotherapy response
We have published a video synopsis of :
Genomic signatures for paclitaxel and gemcitabine resistance in breast cancer derived by machine learning. Stephanie N. Dorman, Katherina Baranova, Joan H.M. Knoll, Brad L. Urquhart, Gabriella Mariani, Maria Luisa Carcangiu, Peter K. Rogan. Molecular Oncology, in press. DOI: http://dx.doi.org/10.1016/j.molonc.2015.07.006
August 28, 2015. Article on chemotherapy gene signature published
The uncorrected proofs of our new paper:
Genomic signatures for paclitaxel and gemcitabine resistance in breast cancer derived by machine learning. Dorman et al. Mol. Oncol. 2015
DOI: 10.1016/j.molonc.2015.07.006
are now available online at this link (http://dx.doi.org/10.1016/j.
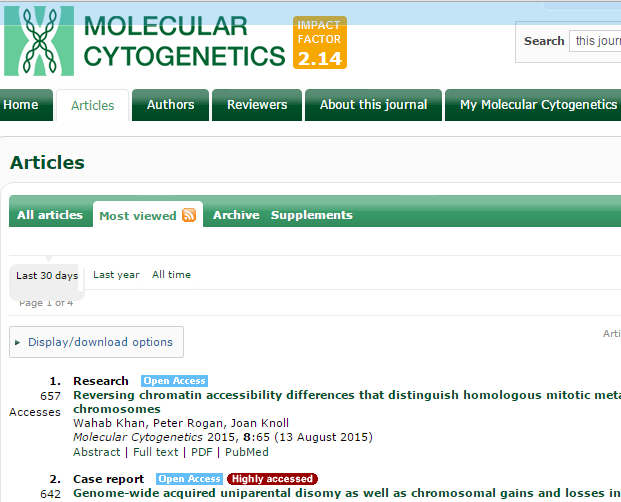
Aug. 25, 2015. Top viewed article in Molecular Cytogenetics
Our article:
Reversing chromatin accessibility differences that distinguish homologous mitotic metaphase chromosomes. Wahab Khan, Peter Rogan, Joan Knoll. Molecular Cytogenetics 2015, 8:65
was published on August 13th. In less than two weeks, it has become the most viewed article in this journal for the past month, averaging 55 per day.
Update: as of Sept. 6, the article is still the top viewed paper with 887 views.
August 13, 2015. New paper on metaphase epigenetics published
Reversing chromatin accessibility differences that distinguish homologous mitotic metaphase chromosomes. Khan et. al. Molecular Cytogenetics 2015, 8:65
July 31, 2015. Chemotherapy resistance in breast cancer manuscript accepted for publication
Genomic signatures for paclitaxel and gemcitabine resistance in breast cancer derived by machine learning
Authors: Stephanie N. Dormana, Katherina Baranovaa, Joan H.M. Knollb,c,d, Brad L. Urquharte, Gabriella Marianif, Maria Luisa Carcangiuf, Peter K. Rogana,d,g,h*
aDepartment of Biochemistry, Schulich School of Medicine and Dentistry, University of Western Ontario, London, ON, Canada, bDepartment of Pathology and Laboratory Medicine, Schulich School of Medicine and Dentistry, University of Western Ontario, London, ON, Canada, cMolecular Diagnostics Division, Laboratory Medicine Program, London Health Sciences Centre, ON, Canada, dCytognomix Inc., London ON, Canada, eDepartment of Physiology and Pharmacology, Schulich School of Medicine and Dentistry, University of Western Ontario, London, ON, Canada, fDepartment of Diagnostic and Laboratory Pathology, Fondazione IRCCS Istituto Nazionale dei Tumori, Milan, Italy, gDepartment of Computer Science, University of Western Ontario, London, ON, Canada, hDepartment of Oncology, Schulich School of Medicine and Dentistry, University of Western Ontario, London, ON, Canada
will be published in a forthcoming issue of the journal “Molecular Oncology”.
July 11, 2015. Presentation at RegGen Satellite Meeting at 2015 ISMB Annual Meeting
We presented:
“Discovery of Primary, Cofactor, and Novel Transcription Factor Binding Site Motifs by Recursive, Thresholded Entropy Minimization”
by Ruipeng Lu, Eliseos Mucaki, and Peter Rogan
at the Regulatory Genomics Special Interest Group meeting in Dublin, Ireland: Link to abstract
July 8, 2015. New paper using Cytognomix’s single copy FISH probes
Khan WA, Rogan PK, Knoll JH. Reversing chromatin accessibility differences that distinguish homologous mitotic metaphase chromosomes. Molecular Cytogenetics, in press.
Stay tuned for posts providing details and links to the manuscript once it is available online at the journal website.
July 3, 2015. New publication on breast cancer gene mutation
FANCM c.5791C>T nonsense mutation (rs144567652) induces exon skipping, affects DNA repair activity, and is a familial breast cancer risk factor.
Peterlongo et al. Hum Mol Genet. 2015 Jun 30. pii: ddv251.
In this paper, we use information theory to demonstrate a new mechanism for disease mutations. It turns out that this a fairly common type of mutation in our unpublished studies of other data sources.
June 27, 2015. Best oral presentation at the 12th Annual London Oncology Research & Education Conference
Natasha Caminsky presented:
A Unified Framework for the Identification and Prioritization of Coding and Non-Coding Variants in Heritable Breast and Ovarian Cancer (HBOC).
which introduces Cytognomix’s approach for analysis of a wide range of regulatory mutations in complete human gene and genome data. The other authors of this study were Mucaki EJ, Lu R, Perri AM, and Rogan PK.
This is an annual event for scientists, clinicians, graduate students and Post-Doctoral fellows at the University of Western Ontario and affiliated hospitals to share cancer research discoveries and promote cancer research collaboration and training. She was awarded Best Oral Presentation for this paper.
The Breast Cancer Society of Canada blogged the results of the paper competition: link
April 18, 2015. New software distribution agreement for MutationForecaster
Today, Cytognomix Inc. and Illumina signed a distribution agreement to make MutationForecaster software available through the BaseSpace ecosystem. Work is underway to enable Illumina users to analyze data processed in BaseSpace to be interpreted with Cytognomix’s software. The BaseSpace environment enables MiSeq users to carry out sequence analyses with requiring an onsite computing infrastructure, with scalable cloud data storage, to manage all of your data analyses, to securely collaborate, and to share and access Illumina and community-developed applications. MutationForecaster will import variant control format files, validate them with RNASeq Bam files from the same sample directly from BaseSpace and download results from Cytognomix back to BaseSpace.
May 21, 2015. Platform presentation at Compute Ontario Research Day
Dr. Peter Rogan’s laboratory at the University of Western Ontario will present:
Discovery of Primary, Cofactor, and Novel Transcription Factor Binding Site Motifs by Recursive, Thresholded Entropy Minimization
by Ruipeng Lu 1, Eliseos Mucaki 2, and Peter Rogan 1,2,3. Departments of (1) Computer Science and (2)Biochemistry, University of Western Ontario, and (3)Cytognomix Inc., London ON
at Compute Ontario (abstract), Conestoga College, Cambridge Ontario, Canada. The presentation is in Room A2107 at 13:55 at the Conestoga College Institute of Technology and Advanced Learning.
May 14, 2015. Presentation at the Great Lakes Chromosome Conference
Dr. Joan Knoll, Chief Scientific Officer of Cytognomix, will present:
Localized, Structural Differences in Condensation of Homologous Metaphase Chromosomes and the Underlying Mechanism
at the 53rd Annual Great Lakes Chromosome Conference at the University of Toronto, Ontario, Canada.
April 30. Sale of MutationForecaster subscription
Cytognomix has sold an annual subscription to MutationForecaster, our comprehensive solution for next generation sequencing based mutation interpretation, to a hospital in Toronto, Ontario Canada. This customer was a previous subscriber to the Automated Site and Exon Definition Server, which is embedded in MutationForecaster and no longer available as standalone software.
April 30, 2015. Sale of Shannon mRNA splicing mutation pipeline software license
Cytognomix’s standalone version of the Shannon mRNA splicing mutation analysis pipeline is distributed through Qiagen CLC bio as a plug-in for their Genomics Workbench and Server software.This product was recently purchased by Dr. Hidetaka Eguchi at the Research Center for Genomic Medicine, Saitama Medical University, Japan.
April 6, 2015. New paper published on cancer of unknown primary
Collaborative effort led by Greg Zaric at the University of Western Ontario:
Identification and survival outcomes of a cohort of patients with cancer of unknown primary in Ontario, Canada. Kim CS1, Hannouf MB, Sarma S, Rodrigues GB, Rogan PK, Mahmud SM, Winquist E, Brackstone M, Zaric GS. Acta Oncol. 2015 Mar 31:1-7. (link)
April 3, 2015. Video tutorials available for MutationForecaster
Videos are now available describing how to use and interpret results from the different components of the MutationForecaster system. Credits: Ben Shirley, Shannon Brown.
Please go to this link to view them all: link
For example, this is a general overview of the system:
March 27, 2015. Cover of the F1000Research webpage
Our paper, “Interpretation of mRNA splicing mutations in genetic disease: review of the literature and guidelines for information-theoretical analysis,” and video, “Interpreting genomic variants in rare and common diseases,” have been highlighted by F1000Research on their home page.