Recently, we published 2 papers describing our unifying framework for non-coding mutation analysis (Mucaki et al. BMC Medical Genomic, 2016; http://bmcmedgenomics.biomedc
(Lu et al. 2016;http://nar.oxfordjournals.org/
I am not claiming that the variants we prioritize with our framework are definitively pathogenic, but do believe that strategies that are narrowly focused on the genetic code itself won’t advance the field or help patients much. Clinical molecular geneticists seriously consider sequencing beyond coding regions and trying to interpret the variants detected in the regions. The incremental costs to do this aren’t exorbitant, and the excuse of ignorance about the meaning of such variants is simply not valid any longer.
Many non-coding mutations have been proven ‘anecdotally’; studies have not been designed to determine the incidence of these types of mutations, in part due to the higher densities of variants in non-coding regions, identifying the clinically relevant ones is more daunting. This has been compounded by the lack of bioinformatic and genomic methods to generate a reliable and comprehensive and high throughput validation of variants outside of coding regions with adverse functional consequences . Suffice it to say, there are many individual reports in the published literature, but they are not generally being systemically uncovered because of the narrow focus on changes in coding regions that affect amino acid sequences.
The problem is not only where the variants reside, but an overly conservative philosophy that fails to consider other interpretations for the effects of variants, even within coding regions. It’s not just non-coding regions that contain missing pathogenic variants, but also coding variants where the change in the amino acid code may not be the source of the disease pathology. There are actually numerous examples of this phenomenon (and a number of good reviews eg. Cartegni et al (https://www.ncbi.nlm.nih.gov/
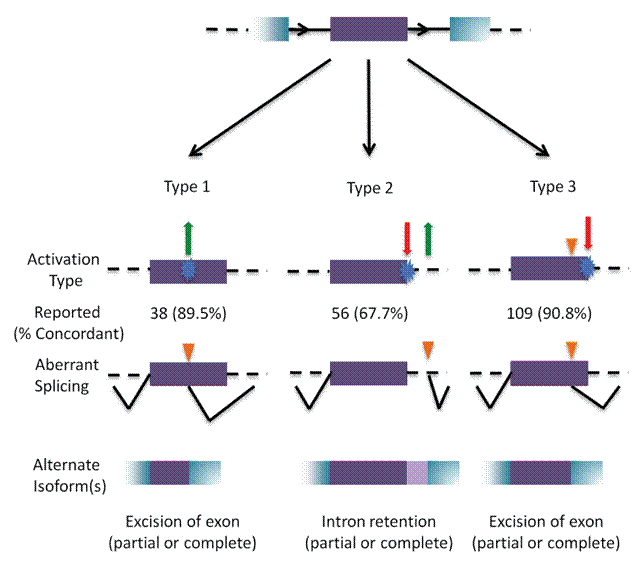
There is inevitably some bias against the reporting of intronic cryptic splicing mutations, because these sequences are not routinely determined in either research or clinical studies. Besides these classes, our studies also identify variants that alter transcription factor binding site strength and mRNA stability (in untranslated regions of mRNAs).
The exchange of mutation information about inherited breast cancer among various testing companies (except Myriad) has increased confidence in mutation interpretation. Those with rare mutations that are not shared among multiple patients do not benefit from this exchange. But these are generally based almost entirely on variants that cause amino acid substitutions or nonsense codons. I contend that such exercises, while very useful, are simply not scalable to the true volumes of all variants found in genes, and they ignore other mechanisms of pathogenicity such as those described above.
To reiterate, my argument is that current clinical molecular diagnostic practices will continue to leave many patients without known pathogenic mutations. Until this point of view changes and we seriously focus on functional and bioinformatic methods to analyze and prioritize VUSs thoughout genes, there will be a lot of frustration about the lack of results among the companies, academics and the patients they are purporting to help. We should also question whether the cost of testing can be justified, with the knowledge that a significant amount of genetic real estate is not being sequenced nor interpreted.
Peter K. Rogan