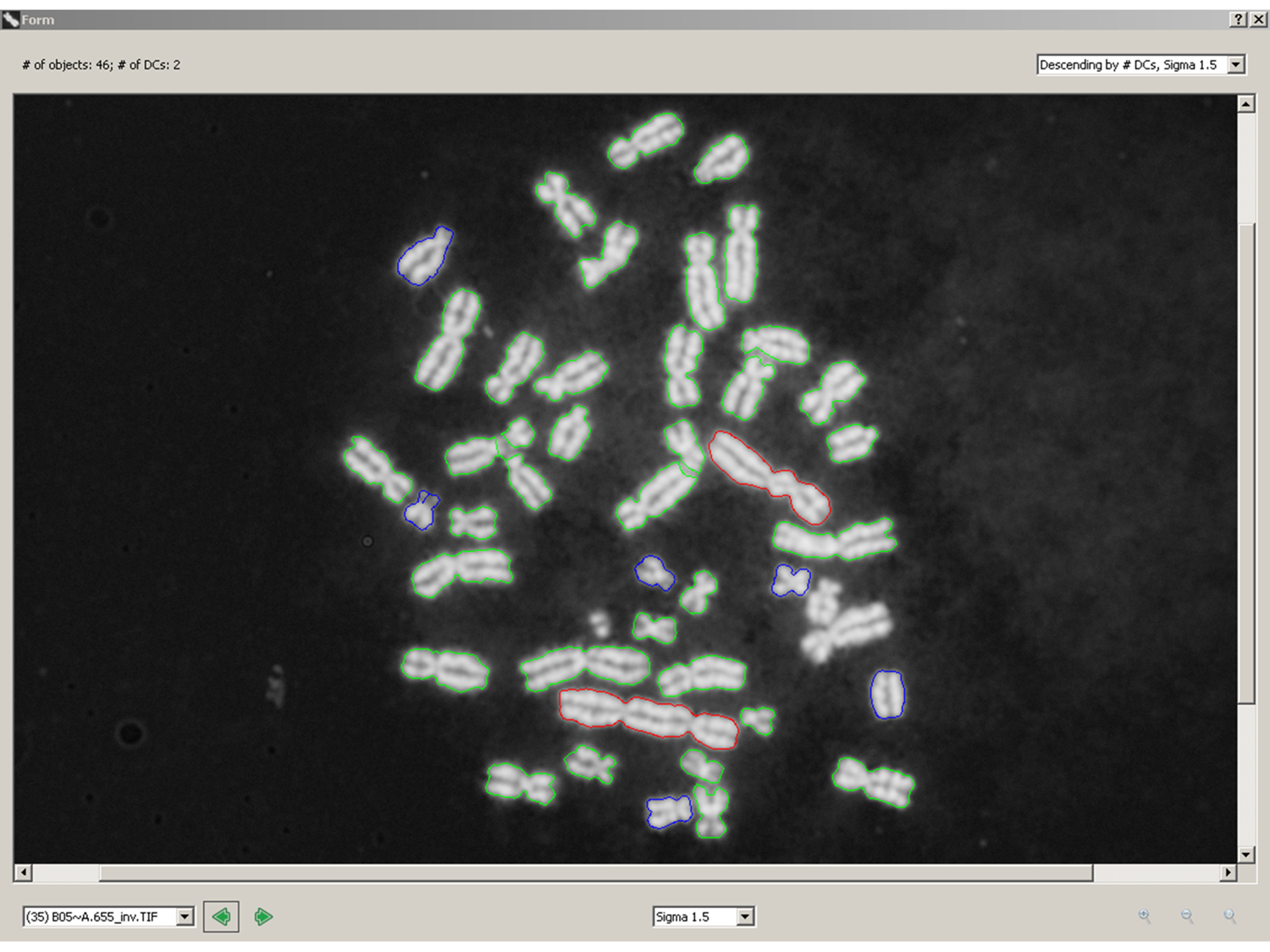
Counting pixel area and pixel intensities (stained antibodies, DNA or RNA) does not determine the identities of the cellular objects that are labeled. The challenge is that every microscope field exhibits different morphology, so traditional image segmentation algorithms aimed at identifying specific subcellular components may not be reliable. We need to be clever to ferret out generalizable image properties of specific cellular components, invariant to morphological variability, that will uniquely discriminate normal from abnormal subcellular distributions of the biomarker of interest. We have done this to identify dicentric chromosomes – see red objects in the attached figure (green are normal, monocentric chromosomes). It should be possible to do this for other subcellular objects. Contact CytoGnomix (mailto://info@cytognomix.com) to discuss further.